Response to Immune Therapy
Project Summary:
For the last several decades lung cancer has been the most common cancer globally and in the US (1). Despite poor patient outcomes associated with diagnosis of lung cancer, the emergence of immunotherapy/immune checkpoint inhibitors have demonstrated durable long-term survival in some patient. However, response rates on checkpoint inhibitor therapies are only ~20% in non-small cell lung cancer (NSCLC) patients and cost of this therapy is high. Immunohistochemistry (IHC) of PD-L1 expression in lung tumor samples is the current modality to determine patient response to immunotherapy. However, there are many limitations of PD-L1 by IHC.
Specifically, approximately 10% of PD-L1- (negative) patients respond to immunotherapy, not all PD-L1+ (positive) patients respond, there is no consensus for IHC positivity for PD-L1, the heterogeneity of PD-L1 expression across different sites of biopsy, the dynamic nature of PD-L1 expression levels that change during the course of therapy and progression, the requirement of tumor specimens, and the antibodies, staining procedures, and quantitation of PD-L1 IHC profile differ among the different companies and different institutions developing anti–PD-1/PD-L1 agents. As such, PD-L1 status alone is a poor biomarker. Complimentary biomarkers are needed to improve prediction of immunotherapy patient response.
Converting digital medical images into high-dimensional data (‘Radiomics’) contains information that reflects underlying pathophysiology and that can be revealed via quantitative analyses (2). Our goal of this study is to build a parsimonious model with high discriminatory capability to predict treatment response in NSCLC patients treated with checkpoint immunotherapy. Baseline only ± delta radiomics (change of features over time) features are being assessed and numerous analytical approaches are being/will be applied including; biostatistical feature reduction analyses, least absolute shrinkage and selection operator, machine learning, neural networks, and deep learning to develop these models.
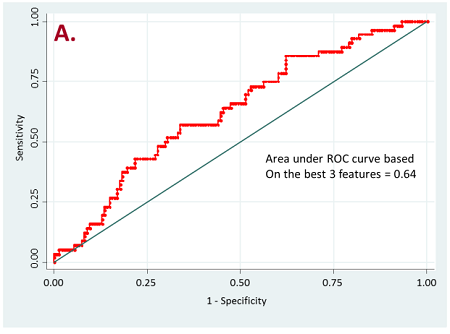
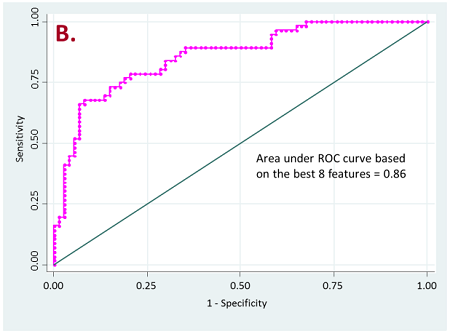
Figure 2. ROC curves for resistant vs long-term responder patients
a) for all target lesions
b) for lung nodules only
Currently, we have retrospectively curated core data elements and CT scans (prior to the initiation of therapy) of 236 patients that have been treated with PD-1, PD-L1, or doublet checkpoint inhibitors. A total of 356 radiomic features based on shape, intensity, texture and wavelets were extracted from the lung tumor region of interest as well as other metastatic sites of each patient. Our initial results have shown that radiomics of lung tumors show better results on discriminating patients that are likely to respond and patients that have resistance to the treatment (Figure 2).
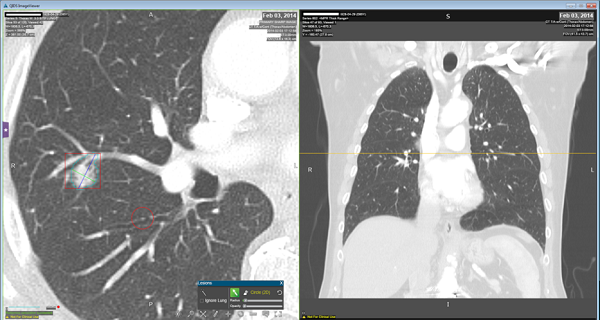
Radiomic features are extracted computationally after the automatic segmentation of the region of interest (i.e. tumor region).

References:
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30.
- Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-6.
Project Members
Robert J. Gillies, PhD
PI, Chair of Department
Matthew B. Schabath, PhD
Co-PI, Associate Member Faculty
Ilke Tunali, MSc
Research Associate
Jhanelle Gray, MD
Thoracic Oncology
Jin Qi, MD
Post-Doc Fellow
Mahmoud Abdallah, PhD
Core Staff Scientist
Yoganand Balagurunathan, PhD
Applied Research Scientist
