BRIGHT
Multimodal Hierarchical Transformers
Asim Waqas
Department of Machine Learning, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department, University of South Florida.
Aakash Tripathi
Department of Machine Learning, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department, University of South Florida.
Ghulam Rasool
Departments of Machine Learning and Neuro-Oncology, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department and Morsani College of Medicine, University of South Florida
(coming soon)
______________________________________________________________________________________________________
Overview
Cancer datasets contain various data modalities recorded at different scales and resolutions in space and time. The goal of this project is to predict cancer recurrence, survival, and distant metastasis for lung, gastrointestinal (GI), and head and neck (HNSCC) cancers using radiological images, pathology slides, molecular data, and electronic health records. Short for Bayesian Hierarchical Graph Neural Networks, BRIGHT models can learn from multimodal, multi-scale, and heterogeneous datasets to predict clinical outcomes in cancer care settings.

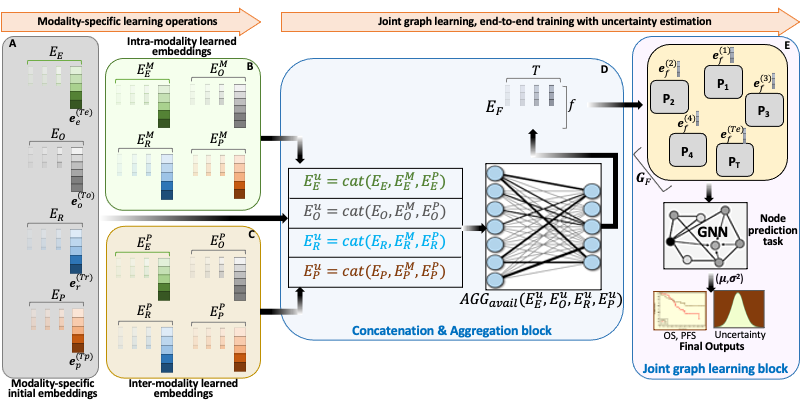
BRIGHT framework for multimodal learning on cancer data. The project involves four learning components (hierarchical, Bayesian, hybrid, and graph structure learning), undertaken in six sequential steps or tasks (pre-training, structure learning, task-specific supervised learning, uncertainty propagation, fine-tuning, and longitudinal retrospective analysis).
______________________________________________________________________________________________________
Method
BRIGHT models address the multimodal learning challenges by combining graph neural networks (GNNs) with other deep learning models using a hierarchical and Bayesian approach, as shown in Fig.6. There are four architectural components of the project that will be undertaken in six sequential tasks. Hierarchical component integrates multi-scale data, the Bayesian framework introduces probability distribution functions over learnable parameters to improve model robustness and explainability, the graph learning handles data heterogeneity, missing samples, and missing modalities challenges, and hybrid learning component uses self-supervised and supervised training scheme to extract and learn contextualized features in the input data. These components will be implemented as a framework consisting of six sequential steps or tasks of the project, given below.
- Pre-training: We will begin with the state-of-the-art architectures for all data modalities of the three cancer types (Lung, HNSCC, GI) and pre-train them on public datasets to generate a preliminary set of embeddings in a shared and unified space. We have already started this self-supervised learning task after transfer learning, using optimal weights for initialization, such as ImageNet weights for radiology/ pathology images.
- Graph-structure learning: This stage involves building graphs using embeddings from the previous task to capture relationships between different modalities of patient data and learn similarities among the patients. GNN takes
the per-modality patient graphs as input and learns the node embeddings, which are then fused with the initial modality embeddings through dot product to generate per-patient modality graphs with improved embeddings. Another GNN then undergoes weakly supervised learning of edge features in modality graphs to estimate patient data structure. - Task-specific supervised learning: The updated patient embeddings are again projected back to patient graphs, and task-specific supervised learning is performed using GNN. A prediction head at the end of GNN outputs the selected clinical outcome using node/sub-graph prediction task.
- Density propagation: This step involves repeating the previous three tasks with an additional learning paradigm involving Bayesian approach. All the neural networks are reconstructed using density propagation approach developed by
PI’s lab, where all mathematical operations involve random variables, and the model output has prediction and uncertainty. Model prioritizes the low variance embeddings by utilizing this uncertainty information during learning that improves the robustness of models. - Fine-tuning: We will fine-tune our framework on three downstream tasks, namely patient survival, cancer recurrence, and distant metastasis, using Moffitt Cancer Center’s internal datasets of Lung, HNSCC, and GI cancers. We will freeze (learnable parameters do not change) parts of the model to facilitate the learning process on internal data.
- Longitudinal analysis: The last task involves analyzing the in-house patient data over temporal dimension to predict prospective clinical outcomes and assist the clinicians in decision making. This longitudinal analysis will consider retrospective medical data of the patients over an elapsed time to predict overall survival, recurrence, and distant metastasis.
______________________________________________________________________________________________________
Results
Modality-Specific Model Selection:
We investigated various modality-specific models for our data modalities, radiology, pathology, -omics, and EHR. Our criterion for evaluating the representativeness or optimality of these modality-specific models includes their predictive performance against the ground-truth data of OS and PFS. As a result of preliminary analysis, we used (1) two flavors of Robust and Efficient MEDical Imaging with Self-supervision (REMEDIS) models for the radiology and histopathology images, (2) GatorTron for the EHR data (including clinical notes, lab tests, and radiology/pathology/surgery reports), and (3) Self-Normalizing Networks (SNNs) for -omics data. Our experiments using pre-trained versions of these models showed that REMEDIS and GatorTron extract optimal embeddings without any fine-tuning/transfer learning for squamous cell carcinoma. On the other hand, SNNs always required fine-tuning, potentially linked to the complexity and variability of the -omics datasets.
Prediction of OS in Lung Cancer Patients:
The preliminary experiments focused on predicting OS for lung squamous cell carcinoma patients using a BRIGHT model and a host of multimodal and uni-modal MLPs. The data were collected at Moffitt (103 patients) and consisted of EHR data (including age at diagnosis, gender, ethnicity, race, smoking status, year of diagnosis, vital status (alive/dead), and tumor cellularity), pathology images, and -omics data (included RNA-Seq expression and protein expression). We generated pathology embeddings using REMEDIS and EHR embeddings using GatorTron, without fine-tuning the models. We trained SNNs for -omics data to generate RNA-Seq expression and protein expression embeddings. We used the same embedding size for all modalities, d=32. Model evaluation was done using C-index and 10-fold cross-validation.
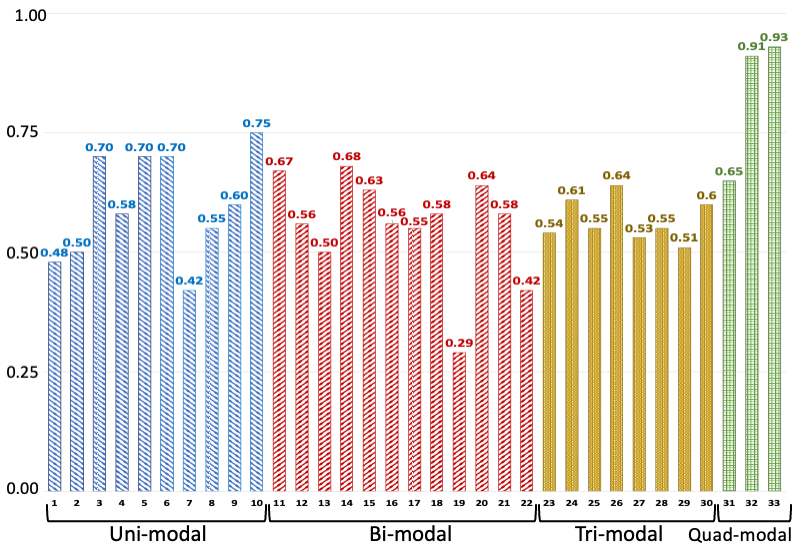
C-index values of various models trained/tested on different combinations of modalities. The legend for the horizontal axis is presented as a tuple: number-(model, modalities): 1-(MLP, EHR), 2-(MLP, Path), 3-(MLP, RNA), 4-(MLP, Protein), 5-(SNN, RNA), 6-(SNN, Protein), 7-(Transfrmr, EHR), 8-(Transfrmr, Path), 9-(Transfrmr, RNA), 10-(Transfrmr, Protein), 11-(MLP, EHR + RNA), 12-(MLP, EHR + Protein), 13-(MLP, EHR + Path), 14-(MLP, RNA + Protein), 15-(MLP, RNA + Path), 16-(MLP, Protein + Path), 17-(Transfrmr, EHR + RNA), 18-(Transfrmr, EHR + Protein), 19-(Transfrmr, EHR + Path), 20-(Transfrmr, RNA + Protein), 21-(Transfrmr, RNA + Path), 22-(Transfrmr, Protein + Path), 23-(MLP, EHR + RNA + Protein), 24-(MLP, EHR + RNA + Path), 25-(MLP, EHR + Protein + Path), 26-(MLP, RNA + Protein + Path), 27-(Transfrmr, EHR + RNA + Protein), 28-(Transfrmr, EHR + RNA + Path), 29-(Transfrmr, EHR + Protein + Path), 30-(Transfrmr, RNA + Protein + Path), 31-(MLP, EHR + RNA + Protein + Path), 32-(Transfrmr, EHR + RNA + Protein + Path), 33-(BRIGHT, EHR + RNA + Protein + Path). We note that BRIGHT models perform better than all other tested models, even the ones that use the same number of modalities, highlighting the importance of graph learning.
______________________________________________________________________________________________________
Conclusion
Convergence of maximum data resolutions across varying time occurrences can accrue remarkable discoveries about the disease indiscernible in individual considerations. BRIGHT project solution aims to converge the entire spectrum of the disease and understand the patient’s genetic, physiological, and psychosocial circumstances in a unified framework. Our framework will help the patient in cancer prevention, early detection, and treatment through informed clinical trials and oncology practices in personalized settings.
______________________________________________________________________________________________________
Citation
@misc{bright-website,
title = {{Bayesian Hierarchical Graph Neural Networks}},
year = {2023},
author = {{Asim Waqas, Aakash Tripathi, Ghulam Rasool}},
note = {Available at: \url{https://lab.moffitt.org/rasool/bright/}}
}
