Pan-cancer Immunotherapy Foundation Models
Foundation Model for Immunotherapy
Aakash Tripathi
Department of Machine Learning, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department, University of South Florida.
Asim Waqas
Department of Machine Learning, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department, University of South Florida.
Ghulam Rasool
Departments of Machine Learning and Neuro-Oncology, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department and Morsani College of Medicine, University of South Florida.
Code
(coming soon)
______________________________________________________________________________________________________
Overview
Immunotherapy using immune checkpoint inhibitors (ICIs) has transformed cancer treatment and improved patient outcomes. Clinical guidelines recommend ICIs based on FDA-approved expression of the checkpoint target, programmed death-ligand 1 (PD-L1), which is measured by immunohistochemistry. Patients who harbor tumors that are PD-L1 positive are associated with statistically significantly higher objective response rates and survival outcomes, clinical responses are widely variable ranging from enduring outcomes and pseudoprogression to rapid progression, hyper-progression, and acquired resistance. We propose to develop a set of adaptable pan-cancer immunotherapy foundation
models (i-FMs).
______________________________________________________________________________________________________
Method
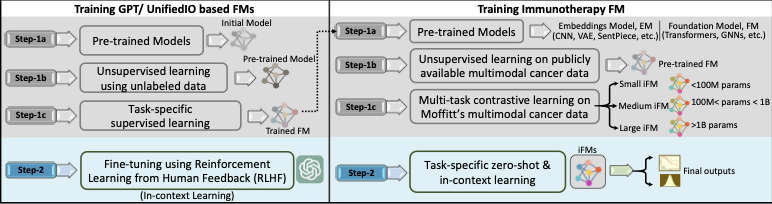
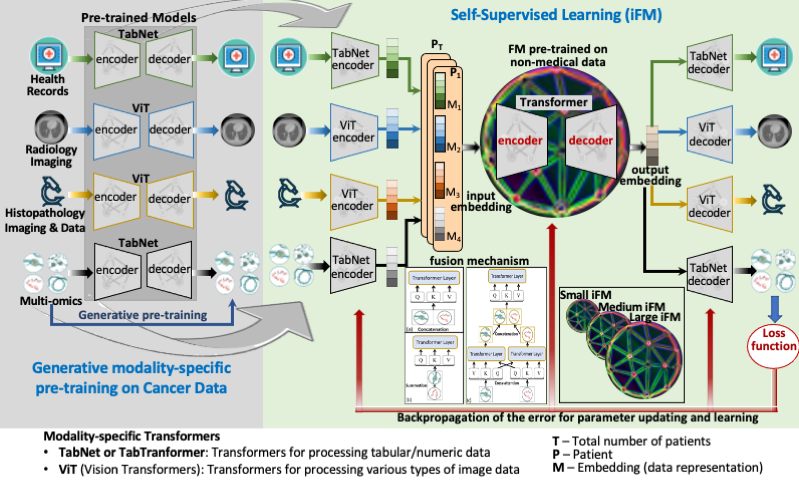
Our hypothesis is that i-FMs can learn immune-related predictive (sub-)visual representations and patterns from multi-modality pan-cancer datasets, and “prompt engineering” using few organ-/disease-specific samples will allow exploiting these learned patterns for accurately predicting survival outcomes. Foundation models (FMs) are initially trained using large unannotated multimodal datasets with self-supervised learning. Once trained, they can be adapted using “prompt engineering” for various downstream tasks with relatively few annotated task-specific examples, much less than required to train a new conventional AI/ML model, such as a convolutional neural network (CNN) or a Transformer. The initial self-supervised training of i-FMs will not require annotated data, i.e., no information about patients’ responses to immunotherapy is needed. Therefore, pan-cancer multimodal data will be used to train three i-FMs (i-FMSMALL with < 100 million parameters, i-FMMEDIUM with 100 million to 1 billion parameters, and i-FMLARGE with 1 billion to 5 billion parameters). Training datasets will include one or more of the following modalities: radiological images, histopathology and immunohistochemistry (IHC) images and data, molecular and other -omics data, and medical records (including demographic information, clinical notes, and lab results, etc.). The final phase of i-FM training will include immune-related data from Moffitt (> 5,700 patients). We will evaluate the trained i-FMs for the downstream task of predicting overall survival (OS) for immunotherapy patients. Prompt engineering will be employed to fine-tune i-FMs using a handful (3 to 5) of annotated examples for three different downstream tasks, i.e., predicting OS for non-small cell lung cancer (NSCLC), head and neck squamous cell carcinoma (HNSCC) and colorectal cancer (CRC). Prompt engineering templates will be created for these three and other cancer sub-types and for longitudinal data that may be available after the start of the immunotherapy. Longitudinal data processing will allow i-FMs to identify patients that may no longer be good candidates for continuing immunotherapy treatment, although they were initially predicted otherwise.
______________________________________________________________________________________________________
Conclusion
The successful completion of the project will result in the development of new pan-cancer biomarkers for immunotherapy using routinely available clinical data and the transformative power of multimodal AI/ML and FMs. Our proposed FM-based biomarkers will extract relevant information from all available clinical data and predict clinical outcomes with high accuracy as compared to the current state-of-the-art biomarkers. FMs are a new class of AI/ML techniques that have the potential to transform current clinical practices due to their ability to learn from very large datasets and provide highly accurate and relevant predictions.
______________________________________________________________________________________________________
Citation
@misc{iFM-website,
title = {{Pan-cancer Immunotherapy Foundation Models}},
year = {2023},
author = {{Asim Waqas, Aakash Tripathi, Ghulam Rasool}},
note = {Available at: \url{https://lab.moffitt.org/rasool/ifm/}}
}
