We are looking for qualified applicants for a post-doctoral fellowship training position in our laboratory. See the description below for ideal candidates:
Postdoctoral Scholar in Cancer Signaling and Proteomics description
In the era of post-genomic medicine, it is important to realize that drug targets in cancer are embedded within a larger system defined by cellular context and this contextual framework has strong influence on the efficacy as well as toxicity of drugs. With the advent of molecular profiling tools such as gene expression profiling, whole genome analysis of copy number variation, and next generation sequencing, we are realizing that cancers can harbor multiple alterations in the genome that enable the ‘hallmarks of cancer’ and these alterations greatly perturb the context of the cancer cell. This system level context therefore is a major driver in the ultimate response to therapy. Alterations in the systems surrounding targets can modulate efficacy in disease and drug targets’ interrelationships with other systems in normal cells can modulate toxicity profiles. The challenge therefore is to annotate such systems surrounding disease targets across a spectrum of disease tissues to best understand molecules that influence effects of primary target inhibition.

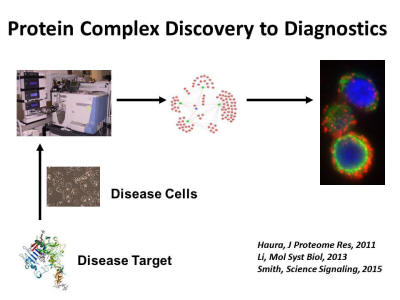
Our approach is to perform deep integrated proteomic studies on cancer to produce more complete views of the tumor architecture allowing contextual understanding of major signaling proteins and drug targets. Alterations in the genomes of cancers ultimately get integrated and produce a cancer proteome that can be analyzed using modern state of the art mass spectrometry proteomic tools. For example, signaling pathways and networks involved in cancers are built using a ‘parts list’ of the cancer genome, such as through integrating mutated genes, genes altered through differential expression and through regulation by micro-RNA molecules. DNA sequencing-based atlases exist for major tumors allowing ‘part lists’ for cancers; however, these atlases lack integration with expressed proteomes and signaling architectures. By taking into account all these alterations in the cancer genome, cancer proteomics can annotate and prioritize proteins and pathways important for cancer growth and survival. Thus, proteomics can view, in sum, small effects of multiple but important mutations in a single cancer. Furthermore, microenvironmental influences, known to be important in drug response, are lacking from these DNA based studies. Proteomics can inform about active pathways driving cancers and lead to novel combination therapy approaches for targeting complex oncogenic networks. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is increasingly used to study cancer proteomes. This includes examining the ‘expressed proteome’ through shotgun proteomics, global signaling by annotating key post-translational events (phosphorylation, acetylation, ubiquination) events in cancers or assembling protein-protein interaction data that yield network views of cancer. This allows unbiased and global views of signaling events in cancer thus offering complementary views of cancer biology that are not considered by sequencing of genes or gene expression. By integrating DNA-RNA-proteome-network type data, the co-existing driver processes instilled by the genome that either surround or act in parallel to drug targets can be mapped directly onto cancer molecular machines that drive cancer progression and response to therapy.
Discovery proteomics has become a widely used tool in our laboratory and we have assembled a diverse team combining clinical medicine, mass spectrometry, chemistry, and bioinformatics/biostatistics to enable deep dive studies into cancer proteomes. This approach provides an unbiased view of the components in a sample, supporting the testing of multiple hypotheses and generating new leads. Our group has made major advances integrating complementary mass spectrometry approaches to build molecular snapshots of cancer proteomes, including expression proteomics in tumors related to drug resistance, drug affinity selection of proteins and identification of drug targets using mass spectrometry, and phosphoproteomics. Our laboratory has also developed approaches to mapping protein-protein interactions or “interactomes” and recently has developed proximity ligation assay methods to translate discovery of protein complexes into in situ assays to annotate protein complexes in human tumor tissues.
Interviews & Press
Adoptive Cell Therapy Plus Checkpoint Inhibitors Show Promise in Non-Small Cell Lung Cancer
Novel Therapy Shows Promise for Lung Cancer Patients with Rare EGFR Mutation
Moffitt Researchers Identify New Relevant Target for PARP Inhibitor Talazoparib
Moffitt Receives $10.2 Million Grant to Develop New Lung Cancer Therapies
Moffitt Researchers Identify Subtypes of Squamous Cell Lung Cancer
The CSBC U01 Research Project @ UCLA/Moffitt/MIT
Science Signaling Podcast for 18 October 2016: ALK combination therapies for lung cancer
New Drug Targets May Boost Protein Inhibitor, Survival for Lung Cancer Patients, Moffitt Cancer Center Study Shows, October 18, 2016
Realistic Postdoc Uses Proteomics To Achieve Base Hits - Moffitt Momentum, Vol 2, Issue 1 (2015)
Moffitt Cancer Center and Ignyta, Inc. Enter into Cancer Research Collaboration for RXDX-106 - Press release, November 20, 2015
Dr. Haura Studies Proteins To Learn About Immune System - Moffitt Momentum, Vol 2, Issue 2 (2015)
Dr. Haura on New Biomarker Assay System for Lung Cancer - OncLive, May 2015
Science Signaling Podcast: 13 January 2015
Interviews with Fox 13 TV on personalized cancer treatment
Behind the Science Video Series - hosted by Eric B. Haura